Joins in tidyverse
Joining two tibbles x and y (like merge in base R)
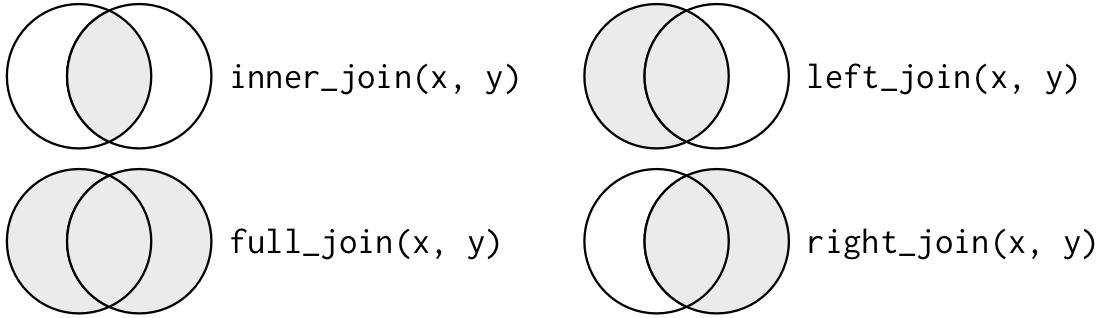
Figure 1: Venn diagram (https://r4ds.had.co.nz/relational-data.html)
Generate some hypothetical indices for some HOBO ids (course 2020):
temp <- tibble(hobo = c(10350086, 10350048, 10347346, 10347319),
t_avg = c(2.3, 3.4, -1.1, 0.8))
temp
# A tibble: 4 x 2
hobo t_avg
<dbl> <dbl>
1 10350086 2.3
2 10350048 3.4
3 10347346 -1.1
4 10347319 0.8Now we want to add the meta information from those HOBOs to the tibble/table.
meta %>% select(-description) %>% as.data.frame()
id hobo_id latitude longitude exposition altitude influence
1 1 10760706 47.9750 7.8260 W 5 low
2 2 10347531 48.0070 7.8212 S 10 moderate
3 3 10760815 48.0139 7.8573 E 3 low
4 4 10350086 47.9907 7.8717 N 3 moderate
5 5 10347319 47.9852 7.8360 S 16 low
6 6 10350043 47.9898 7.8629 N 6 moderate
7 7 10350009 47.9980 7.8372 N 7 low
8 8 10350048 47.9852 7.8885 S 6 low
9 9 10350051 48.0480 7.8120 N 15 high
10 10 10347346 48.0207 7.8101 E 10 low
11 11 10350004 48.0093 7.8170 E 2 moderate
12 12 10350070 48.0106 7.8075 E 3 low
13 13 10350083 47.9944 7.8297 N 6 low
14 14 10610853 47.9797 7.8055 N 1 low
15 15 10347342 47.9892 7.8562 W 3 low
16 16 10347335 48.0057 7.8142 N 3 low
17 17 10347364 48.0265 7.8048 E 1 high
18 18 10347386 47.9965 7.8287 E 1 low
19 19 10347357 48.0175 7.8234 N 4 moderate
20 20 10350099 47.9925 7.8340 W 5 high
21 21 10760763 47.9772 7.8192 S 10 low
22 22 10347337 48.0460 7.5123 W 4 low
23 23 10350090 47.9895 7.8546 E 6 moderate
24 24 10801134 48.0132 7.8483 E 16 high
25 25 10760820 47.9894 7.8415 W 4 low
26 26 10347392 48.0038 7.8544 N 6 moderate
27 27 NA NA NA <NA> NA <NA>
28 28 10350066 48.4105 7.7657 W 3 low
29 29 10347359 48.0031 7.8204 W 15 high
30 30 10350032 47.9851 7.8885 E 1 low
31 31 10350105 47.9905 7.8627 N 10 moderate
32 32 10350084 48.0137 7.8599 S 1 low
33 33 10350007 47.9799 7.8923 E 6 moderate
34 34 10347312 47.9972 7.8307 N 4 moderate
35 35 10347367 47.9747 7.8364 W 1 low
36 36 10234636 48.0191 7.8048 W 2 low
37 37 10350056 47.9926 7.8315 E 3 very high
38 38 10349994 48.0265 7.8048 E 1 high
39 39 10132405 47.9979 7.8373 E 7 moderate
40 40 10347391 47.9987 7.8507 S 4 moderate
41 41 10350049 47.9851 7.8885 E 1 low
42 42 10350029 47.9814 7.8985 E 2 low
43 43 10350053 47.9972 7.8306 S 4 moderate
full_join(temp, meta, by = c("hobo" = "hobo_id"))
# A tibble: 43 x 9
hobo t_avg id latitude longitude exposition altitude influence
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr>
1 1.04e7 2.3 4 48.0 7.87 N 3 moderate
2 1.04e7 3.4 8 48.0 7.89 S 6 low
3 1.03e7 -1.1 10 48.0 7.81 E 10 low
4 1.03e7 0.8 5 48.0 7.84 S 16 low
5 1.08e7 NA 1 48.0 7.83 W 5 low
6 1.03e7 NA 2 48.0 7.82 S 10 moderate
7 1.08e7 NA 3 48.0 7.86 E 3 low
8 1.04e7 NA 6 48.0 7.86 N 6 moderate
9 1.04e7 NA 7 48.0 7.84 N 7 low
10 1.04e7 NA 9 48.0 7.81 N 15 high
# … with 33 more rows, and 1 more variable: description <chr>
left_join(temp, meta, by = c("hobo" = "hobo_id"))
# A tibble: 4 x 9
hobo t_avg id latitude longitude exposition altitude influence
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr>
1 1.04e7 2.3 4 48.0 7.87 N 3 moderate
2 1.04e7 3.4 8 48.0 7.89 S 6 low
3 1.03e7 -1.1 10 48.0 7.81 E 10 low
4 1.03e7 0.8 5 48.0 7.84 S 16 low
# … with 1 more variable: description <chr>Try the same with right_join() and semi_join() and see the differences between the different join commands.
If the column names for joining are identical in both tibbles the by= argument can be dropped. It is also possible to join by more than one variable.
names(temp)[1] <- "hobo_id"
left_join(temp, meta)
# A tibble: 4 x 9
hobo_id t_avg id latitude longitude exposition altitude influence
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl> <chr>
1 1.04e7 2.3 4 48.0 7.87 N 3 moderate
2 1.04e7 3.4 8 48.0 7.89 S 6 low
3 1.03e7 -1.1 10 48.0 7.81 E 10 low
4 1.03e7 0.8 5 48.0 7.84 S 16 low
# … with 1 more variable: description <chr>More on joins: https://r4ds.had.co.nz/relational-data.html#mutating-joins