1. Load the HOBO meta file into R
Load library(tidyverse).
We can use the link given in the github repository overview on https://github.com/data-hydenv/_overview and put following code snippet into R.
data <- read_csv("https://docs.google.com/spreadsheets/d/e/2PACX-1vS_m42YZdulaKJVngJZq51T5CEX36LlqkAnHzOoVmcZXpbHGZ_AofTknbrPGRfWS5PRQ9GygOgkEHdz/pub?output=csv", col_types = cols())
meta <- data %>%
as_tibble() %>%
select(-name_anonym, -description)2. First view on meta table
meta
# A tibble: 43 x 9
id hobo_id data_available region latitude longitude exposition
<dbl> <dbl> <chr> <chr> <dbl> <dbl> <chr>
1 1 1.08e7 yes Freib… 48.0 7.83 W
2 2 1.03e7 yes Freib… 48.0 7.82 S
3 3 1.08e7 no Freib… 48.0 7.86 E
4 4 1.04e7 yes Freib… 48.0 7.87 N
5 5 1.03e7 yes Freib… 48.0 7.84 S
6 6 1.04e7 yes Freib… 48.0 7.86 N
7 7 1.04e7 yes Freib… 48.0 7.84 N
8 8 1.04e7 yes Freib… 48.0 7.89 S
9 9 1.04e7 yes Freib… 48.0 7.81 N
10 10 1.03e7 yes Freib… 48.0 7.81 E
# … with 33 more rows, and 2 more variables: altitude <dbl>,
# influence <chr>
head(meta) %>% as.data.frame()
id hobo_id data_available region latitude longitude exposition
1 1 10760706 yes Freiburg 47.975 7.8260 W
2 2 10347531 yes Freiburg 48.007 7.8212 S
3 3 10760815 no Freiburg 48.014 7.8573 E
4 4 10350086 yes Freiburg 47.991 7.8717 N
5 5 10347319 yes Freiburg 47.985 7.8360 S
6 6 10350043 yes Freiburg 47.990 7.8629 N
altitude influence
1 5 low
2 10 moderate
3 3 low
4 3 moderate
5 16 low
6 6 moderate
meta %>% count(exposition, sort = TRUE)
# A tibble: 4 x 2
exposition n
<chr> <int>
1 E 16
2 N 11
3 W 9
4 S 7
meta %>% count(influence, sort = TRUE) %>%
mutate(relative = n / sum(n))
# A tibble: 4 x 3
influence n relative
<chr> <int> <dbl>
1 low 21 0.488
2 moderate 15 0.349
3 high 6 0.140
4 very high 1 0.0233
ggplot(data = meta , aes(x = altitude)) +
geom_histogram(binwidth = 1, fill = "orange", colour = "black")+
theme_light(20)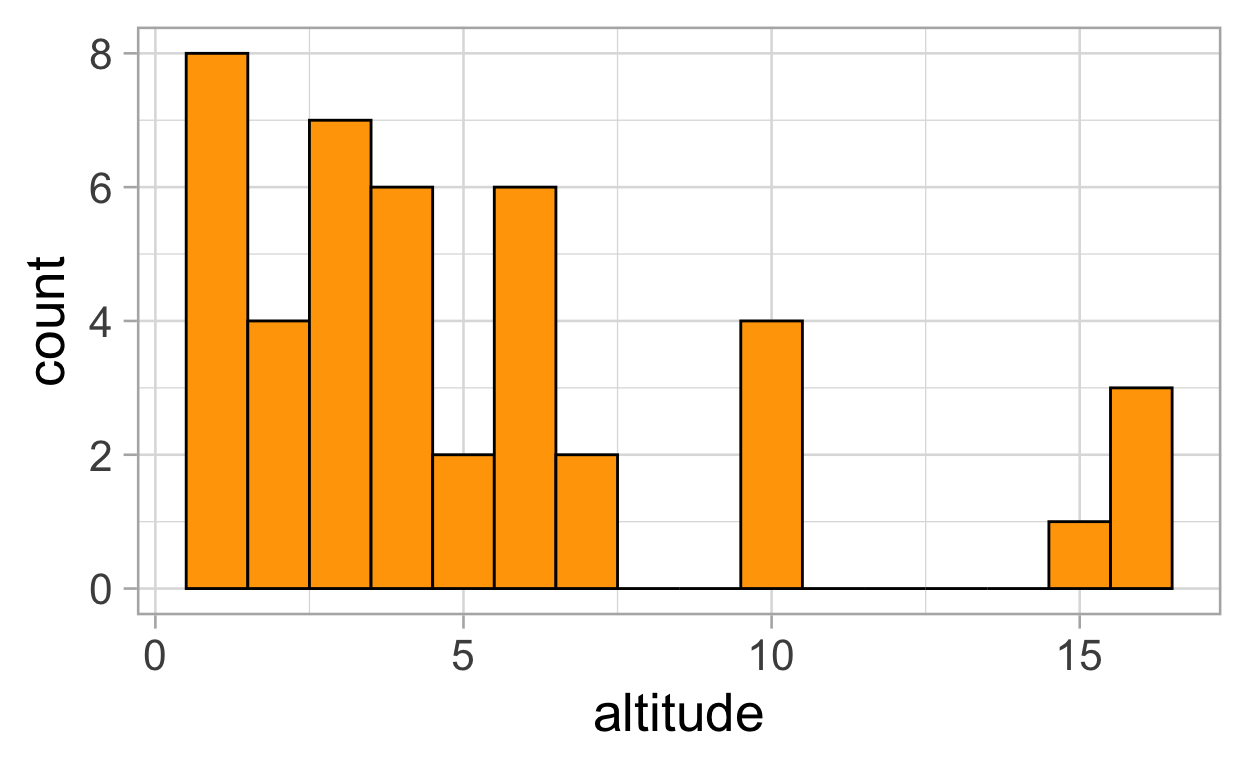
3. Accessing the index sheet
index <- read_csv("https://docs.google.com/spreadsheets/d/e/2PACX-1vS_m42YZdulaKJVngJZq51T5CEX36LlqkAnHzOoVmcZXpbHGZ_AofTknbrPGRfWS5PRQ9GygOgkEHdz/pub?gid=58660777&single=true&output=csv", col_types = cols())
index %>% select(hobo, t_avg, t_fl, f_na, l_md)
# A tibble: 44 x 5
hobo t_avg t_fl f_na l_md
<dbl> <dbl> <dbl> <dbl> <dbl>
1 10760706 5.89 0.525 0.016 1135.
2 10347531 8.76 0.453 0.021 4964.
3 10760815 NA NA NA NA
4 10350086 6.56 0.368 0 90.3
5 10347319 7.65 0.455 0.013 442.
6 10350043 5.83 0.474 0.141 832.
7 10350009 6.72 0.411 0.019 1592.
8 10350048 5.68 0.559 0.172 17495.
9 10350051 NA NA NA NA
10 10347346 6.17 0.362 0.125 6201.
# … with 34 more rowsFor example, then investigating relationships between the indices with a correlation matrix (here from corrr package).
library(corrr)
correlate(index) %>% rplot(shape = 16,
colors = c("magenta","white" ,"blue"))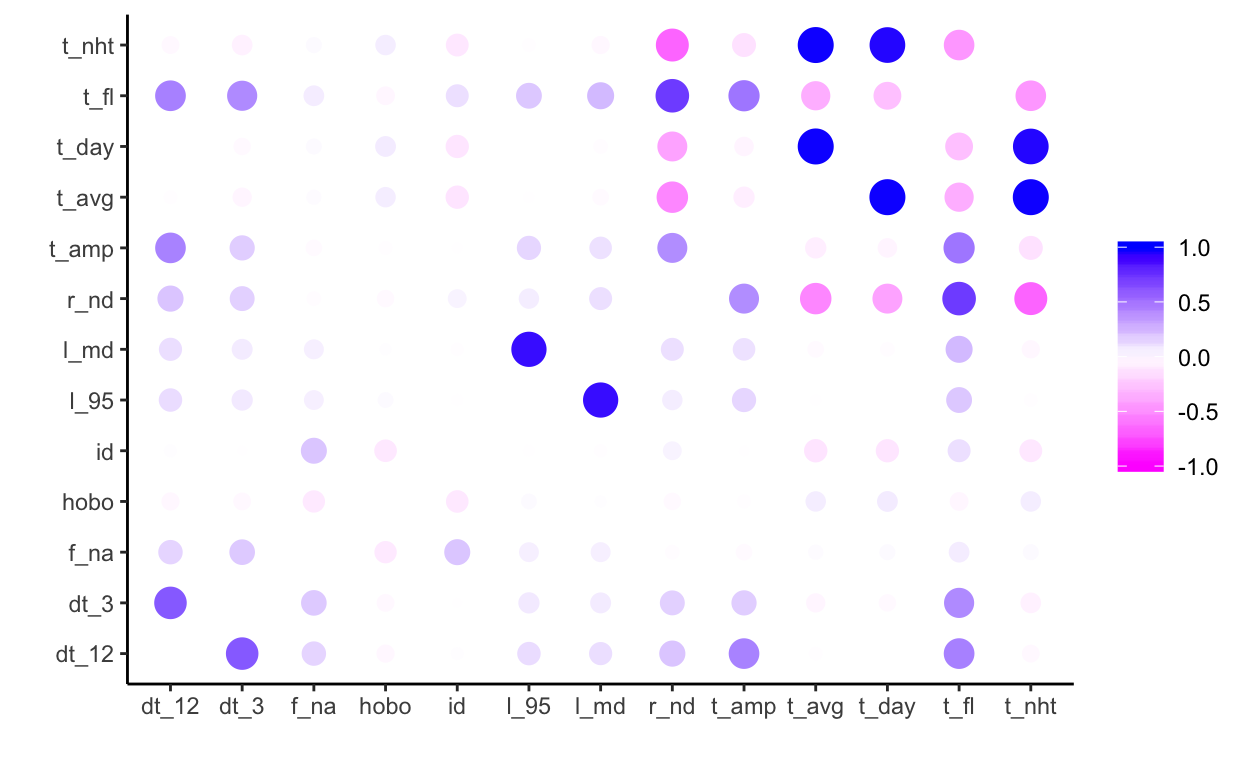
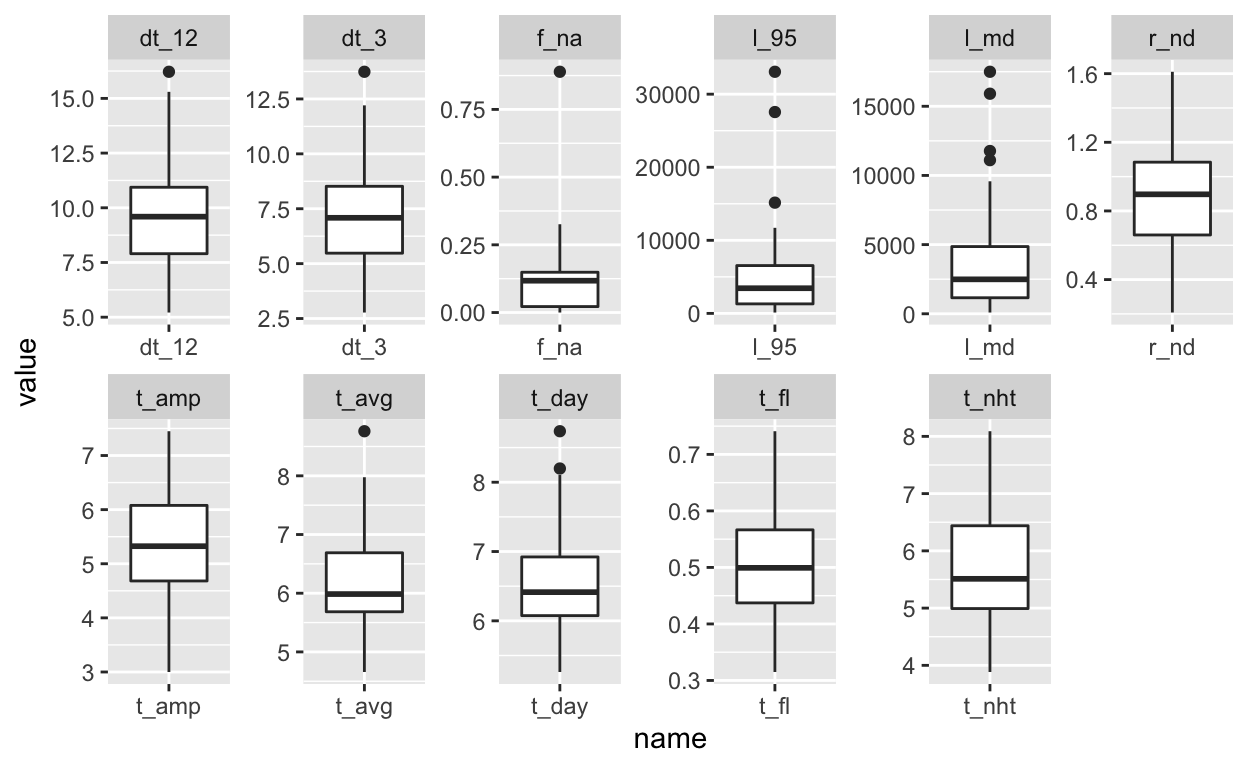
index %>%
pivot_longer(cols = t_avg:l_95) %>%
ggplot(data = ., aes(x = name, y = value))+
geom_boxplot(aes(group=name))+
facet_wrap(~name, scales = "free", nrow = 2)